
- DNA sequencing is the process of determining the entire sequence of nucleotides of a genome.
- The development of new techniques has made it possible to determine the gene sequences of organisms ranging in complexity from bacteria to humans.
- Sanger’s method, also known as dideoxy or chain-termination method was developed by Frederick Sanger in 1977.
- It reduces the DNA replicated from a template strand to be sequenced, into four sets of labeled fragments by interrupting the replication process at one of the four bases.
Principle:
- This method follows the mechanism of DNA replication i.e. DNA synthesis by DNA polymerase.
- The DNA to be sequenced is used as a template strand and a short primer (for the known sequences at 3’ end of the template strand), is radioactively or fluorescently labeled and annealed to it.
- DNA polymerase uses 2’-deoxyribonucleoside triphosphates (dNTPs) as substrates. 3’-hydroxyl group of the primer reacts with the incoming (dNTP) to form a new phospho-diester bond elongating the strand.
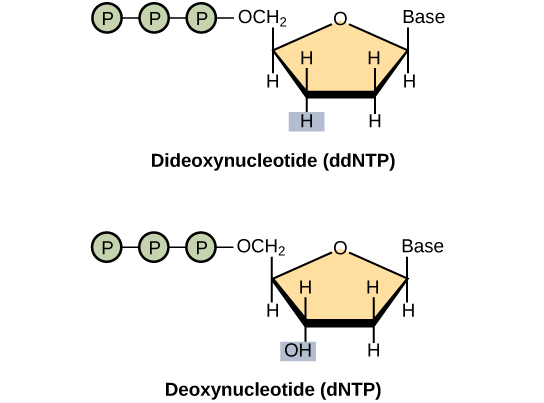
- The reaction mixture also contains modified substrates called 2’-,3’-dideoxynucleoside triphosphate (ddNTPs) which are analogs of dNTPs and lack the 3’-hydroxyl group on their ribose sugar.
https://courses.lumenlearning.com/trident-boundless-microbiology/chapter/bioinformatics/
- Once ddNTP is incorporated at the 3’- end of a growing polynucleotide chain, the lack of 3’-hydroxyl group prevents the addition of further nucleotides through phospho-diester bond causing the elongation to terminate.
- ddNTPS are used in less amount as compared to more amount of dNTPs.
- ddNTPs of respective dNTPs terminate chain at their respective sites. i. e. ddCTP, ddGTP, ddTTP and ddATP terminate at C, G, T and A sites respectively.
- When this process is repeated separately for each of the four ddNTPs, different-sized radiolabeled or fluorescent-labeled nested fragments are obtained. The DNA strands differing in size even by only one nucleotide can be separated.
- Polyacrylamide gel is used for short DNA molecules (up to a few hundred nucleotides) and agarose gel is generally used for longer fragments of DNA.
Procedure:
- Preparation of template for replication:
- The double-stranded DNA is denatured into two single-stranded DNA, taking one as a template DNA.
- A DNA primer which is essential to initiate replication of template is either radiolabeled or fluorescent-labeled and bound with a single stranded cloning vector M13 flanked with the template strand at its 3’-end.
- Many copies of such template DNA hybridized with labeled primer are prepared.
- Generation of four sets of nested labeled fragments:
- Copies of each template are divided into four batches (labeled as A, T, C and G) and each batch is used for different replication reactions.
- DNA polymerase I is added to all four batches containing copies of template.
- Excess of dNTPs substrates (dATP, dGTP, dCTP and dTTP) are added to all the reaction mixtures in each batch.
- Similarly, only one type of ddNTP is added to each respective batch labeled accordingly. i. e. ddATP is added to the batch labeled as A, ddTTP to T, ddCTP to C and ddGTP to G respectively.

https://slideplayer.com/
- Separation of nested DNA fragments by gel electrophoresis:
- The solutions containing a mixture of labeled fragments from four batches are loaded into four different wells on polyacrylamide gel and then electrophoresed.
- DNA being negatively charged, the oligonucleotides will be pulled toward the positive electrode on the opposite side of the gel.
- The smaller a fragment is, the less friction it will experience as it moves through the gel, and the faster it will move. As a result, the oligonucleotides will be arranged from smallest to largest, reading the gel from bottom to top.
- Autoradiogram reading and determination of DNA sequence:
- The autoradiogram of the gel is analyzed to determine the order of bases of complementary strand thus synthesized by DNA polymerase in 5’-3’ direction starting at a provided primer.
- Each terminal ddNTP will correspond to a specific nucleotide in the original sequence. (i. e. the shortest fragment must terminate at the first nucleotide from the 5’ end; the second-shortest fragment must terminate at the second nucleotide from the 5’ end and so on).
- Therefore, by reading the gel bands from smallest to largest, we can determine the 5’ to 3’ sequence of the original DNA strand.