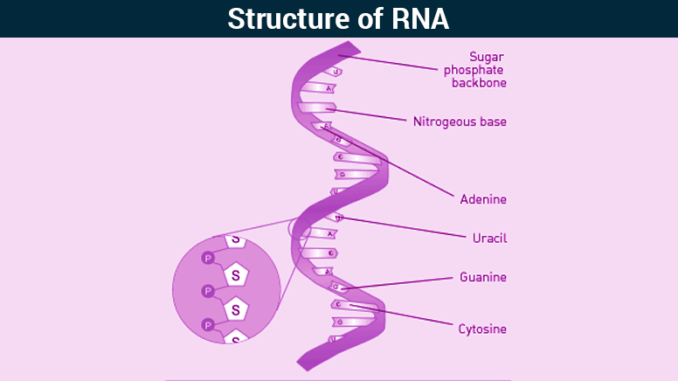
- RNA is a type of nucleic acid found inside living cells.
- It is a polymer of ribonucleotides.
- One molecule of ribonucleotide consists of:
- Ribose sugar
- Nitrogenous base
- Phosphate group
- Nitrogenous bases present in RNA are Adenine, Guanine, Cytosine and Uracil.
- RNA is single-stranded but in most RNA, by means of intra-strand base pairing, they exhibit extensive double-helical character and are capable of folding into diverse tertiary structures.
- Except for the case of certain viruses, RNA is not the genetic material and doesn’t need to be capable of serving as a template for its own replication.
- RNAs are responsible for decoding the information present in the form of genetic code in genes in DNA and also help in protein synthesis.
- There are three types of RNA found in the cell and they are:
- Messenger RNA (mRNA)
- Transfer RNA (tRNA)
- Ribosomal RNA (rRNA)
1. Messenger RNA (mRNA):
- Only a few percent of total cellular RNA is mRNA (only about 5%).
- It is a linear, single-stranded molecule with 5’ and 3’ ends.
- It shows large variations or heterogeneity in its length and nucleotide composition.
- mRNAs contain genetic information for protein synthesis in the form of three-nucleotide codons, which each specify one amino acid.
- Since the message of protein is contained in the codons of mRNA, it is called messenger RNA (mRNA).
- This genetic information is interpreted to generate the linear sequences of amino acids in a polypeptide chain or proteins.
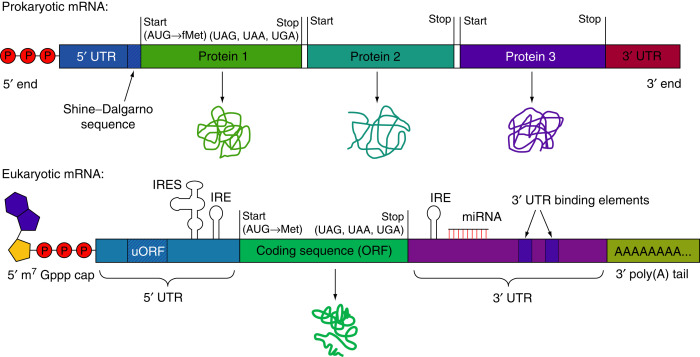
- The protein-coding region(s) of each mRNA is composed of a contiguous, non-overlapping string of codons called an open reading frame (ORF).
- Each ORF specifies a single protein and starts and ends at internal sites within the mRNA. i.e. the ends of an ORF are distinct from the ends of the mRNA.
- Translation starts at the 5’end of the ORF and proceeds one codon at a time to the 3’ end. The first and the last codons of an ORF are known as the start and stop codons.
- Eukaryotic cells always use 5’-AUG-3’ as the start codon whereas in bacteria, the start codon is usually 5’-AUG-3’ but 5’-GUG-3’ and sometimes even 5’-UUG-3’ are also used.
- The start codon has two important functions;
-
-
- First, it specifies the first amino acid to be incorporated into the growing polypeptide chain.
- Second, it defines the reading frame for all subsequent codons.
-
- Stop codons like 5’-UAG-3’, 5’- UGA-3’ and 5’-UAA-3’, define the end of the ORF and signal termination of polypeptide synthesis.
- The number of ORFs per mRNA is different between prokaryotes and eukaryotes.
Source: https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/messenger-rna
- Eukaryotic mRNAs almost always contain a single ORF and hence code for a single polypeptide chain. Such mRNAs are called mono-cistronic mRNAs.
- In contrast, prokaryotic mRNAs frequently contain two or more ORFs and hence can code for multiple polypeptide chains. Such mRNAs containing multiple ORFs are called poly-cistronic mRNAs.
- In eukaryotic cells, 5’-end of mRNA is capped. The 5’ cap is methylated guanine nucleotide (7-methyl guanosine tri-phosphate) that is joined to the 5’ end of mRNA. Similarly, 3’- end is also modified as it has a poly-A tail. Poly-A tail consists of a large number of Adenine nucleotides.
- 5’ cap and poly A tail are absent in prokaryotic cells.
- 5’ cap helps to recognize and bind ribosomes during protein synthesis, whereas poly-A tail enhances the level of translation of the mRNAs by promoting efficient recycling of ribosomes.
- To facilitate binding by a ribosome, many prokaryotic ORFs contain a short sequence upstream (on the 5’ side) of the start codon called the ribosome binding site (RBS) also referred to as Shine-Dalgarno sequence.
Functions of mRNA:
- mRNA acts as a template during protein synthesis. Therefore, it is also known as template RNA. Each codon of mRNA codes for specific amino acid during protein synthesis.
2. Transfer RNA (tRNA):
- It is the smallest type of RNA and accounts for 15% of total RNA in the cell.
- Due to its smallest size, it is also called soluble RNA.
- Basic structure of tRNA of prokaryotes and eukaryotes is same.
- tRNA molecules exhibit a characteristic and highly conserved pattern of single-stranded and double-stranded regions (secondary structure).
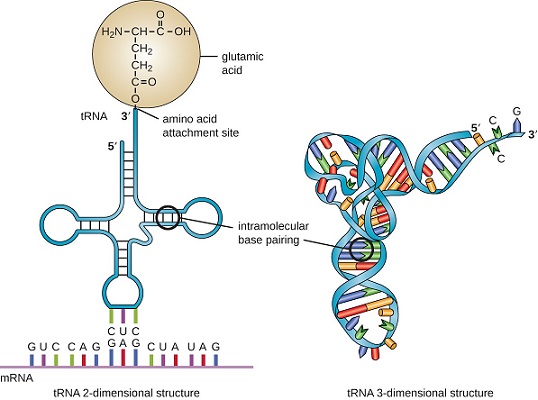
- It is clover-leaf shaped.
- tRNA has four loops where there is no base-pairing whereas in other regions, where bases come closer, they form H-bonds and show a double-stranded structure.
- The principal features of the tRNA cloverleaf are an acceptor stem, three stem loops (referred to as the pseudouridine loop, the D loop and the anticodon loop) and a fourth variable loop.
-
- The acceptor stem is the site of attachment of amino acid that is formed by pairing between the 5’ and 3’ ends of the tRNA molecule. The 5’-CCA-3’ sequence at the extreme 3’ end of the molecule is a single-stranded region that protrudes from this double-stranded stem.
- The pseudo-uridine loop is so named because of the characteristic presence of the unusual base pseudo-uridine (ΨU) in the loop. The modified base is often found within the sequence 5’-T ΨUCG-3’.
- The D loop takes its name from the characteristic presence of dihydrouriidines in the loop.
- The anticodon loop, as its name implies, contains the anticodon, a three-nucleotide- long sequence that is responsible for recognizing the codon by base pairing with the mRNA. The anticodon is always bracketed on the 3’ end by a purine and on its 5’ end by uracil.
- The variable loop sits between the anticodon loop and the ΨU loop and, as its name implies, varies in size from 3 to 21 bases.
- Besides these loops, there are two terminals; 3’-ACC end (which binds amino acid) and a phosphorylated 5’ end.
Functions of tRNA:
- During the process of protein synthesis, charged tRNA transfers activated amino acids into growing polypeptide chain.
- tRNA molecules to which an amino acid is attached are said to be charged and such amino acids attached to a tRNA are called activated amino acids.
- Amino acyl-tRNA synthetases charge tRNAs. Anticodon in tRNA recognizes the codon in mRNA for a specific amino acid. tRNAs are charged by the attachment of that specific amino acid to the 3’- terminal Adenosine nucleotide via high-energy acyl linkage and later on incorporated into a growing polypeptide chain.
3. Ribosomal RNA (rRNA):
- rRNA is the heaviest RNA having high molecular weight (> 1000 KD) and accounts for 80% of the total RNA in a cell.
- It exists in secondary structure in which single strand coils and folds up on itself, however, overall structure of rRNA differs from one another.
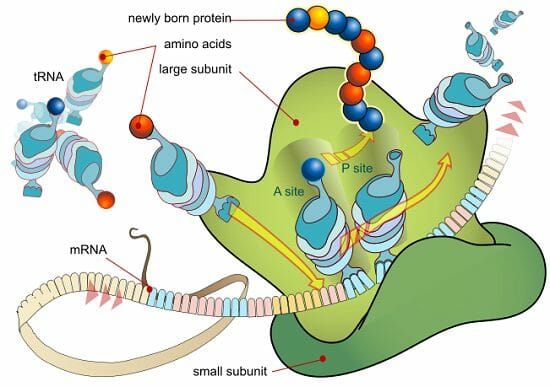
- rRNAs are the structural components of the ribosomes , hence called ribosomal RNA. They are directly responsible for the key functions of the ribosome.
- The ribosome is composed of two subassemblies of RNA and protein known as the large and small subunits.
- The large subunit contains the peptidyl transferase center, which is responsible for the formation of peptide bonds. The small subunit contains the decoding center in which charged tRNAs read or ‘decode’ the codon units of the mRNA.
Source:
https://biologydictionary.net/ribosomal-rna/
- In prokaryotes, 50S and 30S ribosomal subunits unite to form a single intact ribosome of 70S. More than two-thirds of the mass of the prokaryotic ribosomes is RNA. In bacteria, the 50S subunit contains a 5S rRNA and a 23S rRNA, whereas, the 30S subunit contains a single 16S rRNA.
- The eukaryotic ribosome is somewhat larger, composed of 60S and 40S subunits, which together form an 80S ribosome. 40S subunit contains an 18S rRNA whereas, 60S subunit contains a 5S rRNA, 28S rRNA and a 5.8S rRNA (in mammals only).
- The large and small subunits are each composed or one or more rRNAs. 16S and 23S rRNAs are large. On average, a single nucleotide has a molecular mass of 330D; therefore, on its own, the 2900-nucleotide long 23S rRNA has a molecular weight of almost 1000 KD.
- rRNA has a distinctive three-dimensional shape which is made up of internal loops and helices that creates specific sites, A, P, and E, within the ribosome.
Functions of rRNA:
- rRNA provides specific sequence to which an mRNA can bind during the process of protein synthesis. The anticodon loops of the charged tRNAs and the codons of the mRNAs contact the 16S rRNA, not the ribosomal proteins of the smaller subunit during translation.
- It plays a structural role, as in the case of the RNA components of the ribosomal subunits.
- It also acts as enzyme that catalyzes essential reactions in the cell. Such RNAs are also called ribozymes. Hence rRNAs are both structural and catalytic determinants of the ribosome.